Amino acid sequence translation from the nucleotide code of the Pfizer transfection, and an assessment on the presence or lack thereof of HIV insert homology for vaccination end products
*This post was amended on 10/28/2022
The original translation had a slight mistake, which created aberrant ORFs, and made the FCS appear like it was not part of the intended product. This is incorrect, and in reality, the entire Wuhan Hu 1 spike protein is being translated for in the Pfizer vaccine (barring confounding from codon optimization or m1Ψ created read errors). The only differences are again, codon optimization, m1Ψ and the two stabilizing prolines meant to lock in prefusion conformation. There may be alterations to the UTRs as well, or the poly A tail, however I don’t know much about those personally.
https://en.wikipedia.org/wiki/Three_prime_untranslated_region
https://en.wikipedia.org/wiki/Five_prime_untranslated_region
https://en.wikipedia.org/wiki/Polyadenylation
This error occurred because the Ψ symbols in the source document were manually replaced with T’s, to enable its pasting into the translation software. Apologies for the mistake, and the implications are honestly now much worse*
In order to attempt to determine if certain super-antigenic characteristics are maintained in protein translation products created within humans by the Pfizer mRNA “vaccines”, a translation was done of the on record nucleotide code for the Pfizer mRNA LNP based transfection. The source paper this information was located in, as well as the actual download link for the sequence, are listed below:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8310186/pdf/vaccines-09-00734.pdf
https://web.archive.org/web/20210105162941/https://mednet-communities.net/inn/db/media/docs/11889.doc
I should specify by stating, it would be much more likely that vaccine expressed spike, had high affinity for ACE 2, if certain regions were proven to still have highly similar amino acid orders as the original strain. Codon optimization and m1Ψ https://en.wikipedia.org/wiki/N1-Methylpseudouridine
are confounders, and I will not be assessing regions other than the proposed HIV inserts, so this is far from conclusive evidence of high ACE2 affinity for transfection end products. However, showing those regions gone in the final product, would be reason to suspect low affinity or reduced affinity, as they are the sections that deviate from the original SARS the most due to their sheer length, meaning they are most likely to be the regions that confer increased ACE 2 affinity. They are also the regions that contain the furin cleavage site, so without its presence, vaccine created spike may not be capable of being split into S1 and S2 subunit fragments after binding to the ACE 2 receptor, reducing circulating S1 subunits. Thus, it may also reduce the likelihood that ADAM17 accidentally cleaves ACE 2 bound spike while attempting to create sACE2, creating circulating sACE 2 viral complexes. In contrast still seeing them still present, is an indicator that ACE2 affinity may still be high, and thus RAAS dysregulation via ACE2 downregulation, may be likely in vaccination end products. If that were true, this pathway could be a large contributor to vaccine induced injury, thanks to the inflammatory cascades it causes (depicted below)
https://europepmc.org/backend/ptpmcrender.fcgi?accid=PMC8099789&blobtype=pdf
From the above literature, the following sequence, was placed in the following translation software:
https://web.expasy.org/translate/
GAGAATAAAC TAGTATTCTT CTGGTCCCCA CAGACTCAGA GAGAACCCGC
CACCATGTTC GTGTTCCTGG TGCTGCTGCC TCTGGTGTCC AGCCAGTGTG
TGAACCTGAC CACCAGAACA CAGCTGCCTC CAGCCTACAC CAACAGCTTT
ACCAGAGGCG TGTACTACCC CGACAAGGTG TTCAGATCCA GCGTGCTGCA
CTCTACCCAG GACCTGTTCC TGCCTTTCTT CAGCAACGTG ACCTGGTTCC
ACGCCATCCA CGTGTCCGGC ACCAATGGCA CCAAGAGATT CGACAACCCC
GTGCTGCCCT TCAACGACGG GGTGTACTTT GCCAGCACCG AGAAGTCCAA
CATCATCAGA GGCTGGATCT TCGGCACCAC ACTGGACAGC AAGACCCAGA
GCCTGCTGAT CGTGAACAAC GCCACCAACG TGGTCATCAA AGTGTGCGAG
TTCCAGTTCT GCAACGACCC CTTCCTGGGC GTCTACTACC ACAAGAACAA
CAAGAGCTGG ATGGAAAGCG AGTTCCGGGT GTACAGCAGC GCCAACAACT
GCACCTTCGA GTACGTGTCC CAGCCTTTCC TGATGGACCT GGAAGGCAAG
CAGGGCAACT TCAAGAACCT GCGCGAGTTC GTGTTTAAGA ACATCGACGG
CTACTTCAAG ATCTACAGCA AGCACACCCC TATCAACCTC GTGCGGGATC
TGCCTCAGGG CTTCTCTGCT CTGGAACCCC TGGTGGATCT GCCCATCGGC
ATCAACATCA CCCGGTTTCA GACACTGCTG GCCCTGCACA GAAGCTACCT
GACACCTGGC GATAGCAGCA GCGGATGGAC AGCTGGTGCC GCCGCTTACT
ATGTGGGCTA CCTGCAGCCT AGAACCTTCC TGCTGAAGTA CAACGAGAAC
GGCACCATCA CCGACGCCGT GGATTGTGCT CTGGATCCTC TGAGCGAGAC
AAAGTGCACC CTGAAGTCCT TCACCGTGGA AAAGGGCATC TACCAGACCA
GCAACTTCCG GGTGCAGCCC ACCGAATCCA TCGTGCGGTT CCCCAATATC
ACCAATCTGT GCCCCTTCGG CGAGGTGTTC AATGCCACCA GATTCGCCTC
TGTGTACGCC TGGAACCGGA AGCGGATCAG CAATTGCGTG GCCGACTACT
CCGTGCTGTA CAACTCCGCC AGCTTCAGCA CCTTCAAGTG CTACGGCGTG
TCCCCTACCA AGCTGAACGA CCTGTGCTTC ACAAACGTGT ACGCCGACAG
CTTCGTGATC CGGGGAGATG AAGTGCGGCA GATTGCCCCT GGACAGACAG
GCAAGATCGC CGACTACAAC TACAAGCTGC CCGACGACTT CACCGGCTGT
GTGATTGCCT GGAACAGCAA CAACCTGGAC TCCAAAGTCG GCGGCAACTA
CAATTACCTG TACCGGCTGT TCCGGAAGTC CAATCTGAAG CCCTTCGAGC
GGGACATCTC CACCGAGATC TATCAGGCCG GCAGCACCCC TTGTAACGGC
GTGGAAGGCT TCAACTGCTA CTTCCCACTG CAGTCCTACG GCTTTCAGCC
CACAAATGGC GTGGGCTATC AGCCCTACAG AGTGGTGGTG CTGAGCTTCG
AACTGCTGCA TGCCCCTGCC ACAGTGTGCG GCCCTAAGAA AAGCACCAAT
CTCGTGAAGA ACAAATGCGT GAACTTCAAC TTCAACGGCC TGACCGGCAC
CGGCGTGCTG ACAGAGAGCA ACAAGAAGTT CCTGCCATTC CAGCAGTTTG
GCCGGGATAT CGCCGATACC ACAGACGCCG TTAGAGATCC CCAGACACTG
GAAATCCTGG ACATCACCCC TTGCAGCTTC GGCGGAGTGT CTGTGATCAC
CCCTGGCACC AACACCAGCA ATCAGGTGGC AGTGCTGTAC CAGGACGTGA
ACTGTACCGA AGTGCCCGTG GCCATTCACG CCGATCAGCT GACACCTACA
TGGCGGGTGT ACTCCACCGG CAGCAATGTG TTTCAGACCA GAGCCGGCTG
TCTGATCGGA GCCGAGCACG TGAACAATAG CTACGAGTGC GACATCCCCA
TCGGCGCTGG AATCTGCGCC AGCTACCAGA CACAGACAAA CAGCCCTCGG
AGAGCCAGAA GCGTGGCCAG CCAGAGCATC ATTGCCTACA CAATGTCTCT
GGGCGCCGAG AACAGCGTGG CCTACTCCAA CAACTCTATC GCTATCCCCA
CCAACTTCAC CATCAGCGTG ACCACAGAGA TCCTGCCTGT GTCCATGACC
AAGACCAGCG TGGACTGCAC CATGTACATC TGCGGCGATT CCACCGAGTG
CTCCAACCTG CTGCTGCAGT ACGGCAGCTT CTGCACCCAG CTGAATAGAG
CCCTGACAGG GATCGCCGTG GAACAGGACA AGAACACCCA AGAGGTGTTC
GCCCAAGTGA AGCAGATCTA CAAGACCCCT CCTATCAAGG ACTTCGGCGG
CTTCAATTTC AGCCAGATTC TGCCCGATCC TAGCAAGCCC AGCAAGCGGA
GCTTCATCGA GGACCTGCTG TTCAACAAAG TGACACTGGC CGACGCCGGC
TTCATCAAGC AGTATGGCGA TTGTCTGGGC GACATTGCCG CCAGGGATCT
GATTTGCGCC CAGAAGTTTA ACGGACTGAC AGTGCTGCCT CCTCTGCTGA
CCGATGAGAT GATCGCCCAG TACACATCTG CCCTGCTGGC CGGCACAATC
ACAAGCGGCT GGACATTTGG AGCAGGCGCC GCTCTGCAGA TCCCCTTTGC
TATGCAGATG GCCTACCGGT TCAACGGCAT CGGAGTGACC CAGAATGTGC
TGTACGAGAA CCAGAAGCTG ATCGCCAACC AGTTCAACAG CGCCATCGGC
AAGATCCAGG ACAGCCTGAG CAGCACAGCA AGCGCCCTGG GAAAGCTGCA
GGACGTGGTC AACCAGAATG CCCAGGCACT GAACACCCTG GTCAAGCAGC
TGTCCTCCAA CTTCGGCGCC ATCAGCTCTG TGCTGAACGA TATCCTGAGC
AGACTGGACC CTCCTGAGGC CGAGGTGCAG ATCGACAGAC TGATCACAGG
CAGACTGCAG AGCCTCCAGA CATACGTGAC CCAGCAGCTG ATCAGAGCCG
CCGAGATTAG AGCCTCTGCC AATCTGGCCG CCACCAAGAT GTCTGAGTGT
GTGCTGGGCC AGAGCAAGAG AGTGGACTTT TGCGGCAAGG GCTACCACCT
GATGAGCTTC CCTCAGTCTG CCCCTCACGG CGTGGTGTTT CTGCACGTGA
CATATGTGCC CGCTCAAGAG AAGAATTTCA CCACCGCTCC AGCCATCTGC
CACGACGGCA AAGCCCACTT TCCTAGAGAA GGCGTGTTCG TGTCCAACGG
CACCCATTGG TTCGTGACAC AGCGGAACTT CTACGAGCCC CAGATCATCA
CCACCGACAA CACCTTCGTG TCTGGCAACT GCGACGTCGT GATCGGCATT
GTGAACAATA CCGTGTACGA CCCTCTGCAG CCCGAGCTGG ACAGCTTCAA
AGAGGAACTG GACAAGTACT TTAAGAACCA CACAAGCCCC GACGTGGACC
TGGGCGATAT CAGCGGAATC AATGCCAGCG TCGTGAACAT CCAGAAAGAG
ATCGACCGGC TGAACGAGGT GGCCAAGAAT CTGAACGAGA GCCTGATCGA
CCTGCAAGAA CTGGGGAAGT ACGAGCAGTA CATCAAGTGG CCCTGGTACA
TCTGGCTGGG CTTTATCGCC GGACTGATTG CCATCGTGAT GGTCACAATC
ATGCTGTGTT GCATGACCAG CTGCTGTAGC TGCCTGAAGG GCTGTTGTAG
CTGTGGCAGC TGCTGCAAGT TCGACGAGGA CGATTCTGAG CCCGTGCTGA
AGGGCGTGAA ACTGCACTAC ACATGATGAC TCGAGCTGGT ACTGCATGCA
CGCAATGCTA GCTGCCCCTT TCCCGTCCTG GGTACCCCGA GTCTCCCCCG
ACCTCGGGTC CCAGGTATGC TCCCACCTCC ACCTGCCCCA CTCACCACCT
CTGCTAGTTC CAGACACCTC CCAAGCACGC AGCAATGCAG CTCAAAACGC
TTAGCCTAGC CACACCCCCA CGGGAAACAG CAGTGATTAA CCTTTAGCAA
TAAACGAAAG TTTAACTAAG CTATACTAAC CCCAGGGTTG GTCAATTTCG
TGCCAGCCAC ACCCTGGAGC TAGCAAAAAA AAAAAAAAAA AAAAAAAAAA
AAAAGCATAT GACTAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA
AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAA
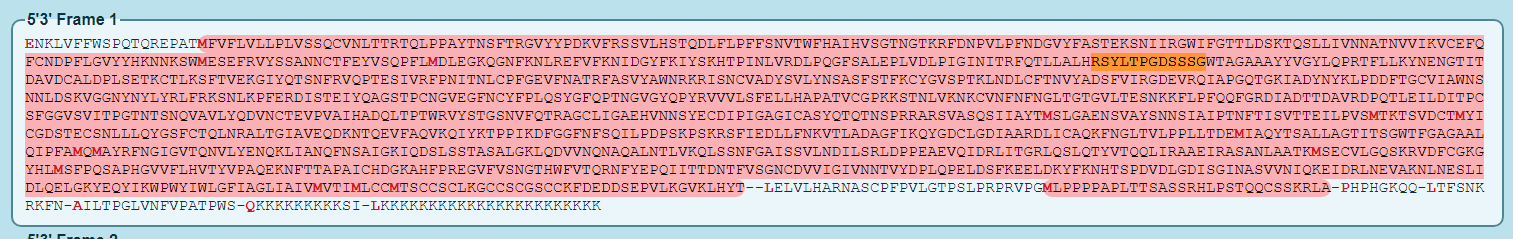
In the event first reading is abberhant, protein production is initiated from a subsequent reading frame, and the translation product may be changed. These are known as open reading frames. It is most likely, that the intended product for this sequence is ORF1 in the 5’3 direction, as that is the intended read direction as well as what is likely the intended start codon.
As you can see, the entire spike protein is being created in the intended product by ORF1 5’3 (keep in mind the other open reading frames may still occur in error.) The protein produced is identical to the Wuhan Hu spike sequence (except for two stabilizing prolines, altered to attempt to lock the spike into prefusion confirmation)
https://www.genome.jp/dbget-bin/www_bget?refseq:NC_045512
Next, I control F to search the ORFs for sections of the known HIV epitopes from multiple preprints (covered in the previous article), depicted below:
The spike being identical, is honestly, shocking. 4/4 of the inserts that have HIV homology are still present, and they have not been attenuated in any way. the two stabilizing Ps are the only notable change between the Wuhan Hu 1 spike protein and the spike protein that is supposed to be produced by the vax (again, Codon optimization and m1Ψ https://en.wikipedia.org/wiki/N1-Methylpseudouridine
are confounders)
As I stated at the time, this is a rather bad sign. If I could show these regions gone, I could feasibly assume that vaccine created spike proteins would have less affinity for ACE 2 receptors. However the only alteration I can find (from the first paper linked) is the two prolines for prefusion locking. Prefusion would be in the state the spike is in before undergoing membrane fusion. This is also not likely reducing ACE2 affinity, as prefusion is the state spike is in before binding occurs, meaning it is the state most likely to have an electromagnetic attraction to ACE 2, since it’s RBD is exposed to attempt binding in this state.
Final verdict: 4/4 HIV inserts present in intended product, FCS still present and not crippled in the slightest. Likelihood of maintained ACE 2 receptor affinity is thus high
This also means, that if the preprint from the previous article is correct, and the FCS can in some cases double as a nuclear localization sequence, that reverse transcription may be a plausible concern with the vaccination. Remember the FCS has mutated in Omicron, (check previous article covering NLS)
*THOUGHTS: Is NLS actually not indicative of reverse transcription? Can produced proteins be reversed transcribed, or only nucleic acids? Does an NLS in the RNA genome play any other role? Actually may be implications for retracted VDJ recombination paper:
https://www.mdpi.com/1999-4915/13/10/2056/htm
If an error in interpreting this material was made, feel free to leave a comment illustrating how.