A hypothetical biological pathway that may link SARS CoV 2 vaccinations to giant cell pathologies, via CD4 derangement due to receptor binding of HIV GP 120 homologous regions, Part 2
To continue to provide evidence, I would now like to shift away from protein structure and instead ponder on certain possibilities, to attempt to determine if the prior proposals hold merit. To make it more clear, I propose that the HIV glycoprotein homology in the N and C terminal domains of the SARS CoV 2 spike S1 subunit, may cause said spike to bind to host immune receptors, specifically those present on CD4 T cells1. I also propose this to be a much greater risk in the vaccination end products than I do for most viral infections, due to innate barrier2 bypass (mucus, blood air barrier3, secretory IgA4 ect), and deleterious quasiswarm5 mutations, that will result in improperly packaged replication incompetent virions and fragments of virions. Codon optimization67 may also play a role in the LNP platforms as well, but for now, I’ll stop the postulations concerning replication count there, as I feel bypassing already discussed evolutionarily established viral barriers, and only choosing to replicate the spike protein, pretty much assures a greater production of GP120 epitopes in a vaccinated individual, than likely the most acutely infected ones. The infected ones will also give said epitopes much less/restricted access to their circulatory system, which is where I feel the majority of damage may be done by deranged CD4 cells.
We will start by looking at each individual receptor that GP120 normally interacts with, then we shall assess real world data, to see if our hypothesis has the potential to hold up to scrutiny. The information pertaining to HIV and receptor interaction can be found below8:
”The gp120s are exposed for the sequential and specific recognition of two membrane proteins of the host cell, the receptor CD4 and a chemokine receptor acting as coreceptor, either CC chemokine receptor type 5 (CCR5) or CXC chemokine receptor 4 (CXCR4). The binding of gp120 to cellular receptors brings the virus closer to the host cell and leads to exposure of gp41 that inserts into the host cell plasma membrane.”
CCR5:
C-C chemokine9 receptor type 5, aka CCR5 or sometimes CD195, is a protein receptor for chemokines. It’s a seven trans-membrane G-protein10 coupled receptor, which mediates physiological functions of many types of immune cells11 (T cells, macrophages, eosinophilis, meyloid derived suppressor cells, microglia, and dendritic cells)
Depicted below is how exactly GP120 interacts with CCR5 in normal HIV infection. Let’s also go over, how this passage is evidence AGAINST our hypothesis, before we continue to cover evidence that corroborates our theory
“Binding to CD4 stabilizes an open conformation of Env (the gp160 complex), which likely preexists in equilibrium with the closed one [9] and other open conformations [10]. Opening of the trimer involves outward movement and rotation of gp120 subunits, while the C-terminal part of gp41s HR1 domain elongates to adopt a fusion-competent conformation. The gp120s V1/V2 loops move from the trimer apex to the sides of the trimer, releasing the V3 loop, which contains determinants for binding the coreceptor. At the boundary between gp120’s inner and outer domains, movement of V1/V2 loops also triggers formation of the four-stranded bridging sheet domain (BS), which also contributes to coreceptor binding”
and this photo and passage, which show us the exact variable loops responsible for said receptor interaction. This is the point which may be evidence against us
*AMENDMENT: THE “BRIDGING SHEET” IS COMPOSED OF VARIABLE LOOPS 1 AND 2. WHILE V LOOP 3 NOT BEING IN THE INSERTS IS EVIDENCE AGAINST US, AS WILL BE MOMENTARILY COVERED, I DID NOT REALIZE AT THE TIME OF WRITING, THAT THE BRIDGING SHEET IS COMPOSED PARTIALLY OF VARIABLE LOOPS STEMS 1 AND 212
”The bridging sheet is a four-stranded, antiparallel b sheet that includes the V1/V2 stem and strands (b20 and b21) derived from the fourth conserved gp120 region. CD4 contacts gp120 residues in the outer domain and the bridging sheet (6). The gp120 residues implicated by our study in CCR5 binding are located near or within the bridging sheet”
INSERT 3 IS FOUND IN VARIABLE LOOP 1 OF THE PRADHAN PAPER, PROVIDED THEIR ALIGNMENT IS CORRECT. IF NOT, INSERT 3 BEARS HIGH HOMOLOGY TO THE ON RECORD CD4 BINDING LOOP, HOWEVER THIS PARTICULAR ARTICLE IS ABOUT CCR5, SO WE WILL IGNORE THAT DATA UNTIL WE FULLY COVER THE CD4 RECEPTOR, IN A FUTURE ARTICLE
AMENDMENT CONCLUDED.
As we can see, variable loop 3 is the one (along with the bridging sheet) that is supposedly responsible for CCR5 binding in this assessment. V loops 1 and 2 change confirmation, opening up to allow V loop 3 to facilitate this interaction. If we look again at the Pradhan alignment summary, we can see that variable 3, is not where any of the inserts lie. On the contrary, they lie in V1, V5, and V4
We now once again need to note, that these are variable loops, and they are aptly named. They are highly mutative, and also possess extensive glycosylation, to aid with their ability to evade patrolling immune cells or slip antibody binding. We also need to remember, that the loops above are from variants of HIV, spread across the globe, and that all humans are different. For example, a known mutation in this receptor (CCR5-Δ32) is:
“a 32-base-pair deletion that introduces a premature stop codon into the CCR5 receptor locus, resulting in a nonfunctional receptor.[43][44] CCR5 is required for M-tropic HIV-1 virus entry.[45] Individuals homozygous (denoted Δ32/Δ32) for CCR5 Δ32 do not express functional CCR5 receptors on their cell surfaces and are resistant to HIV-1 infection, despite multiple high-risk exposures.[45]”13
I posit, that other variable loops other than 3 may well have an attraction or functional interaction with CCR5, that has either yet to be discovered, or possibly arose over time as HIV mutated, and gave rise to these variants. It is also possible, that CCR5 receptor interaction in the context of SARS 2 is created or facilitated by still unknown differences in the SARS 2 spike versus the GP160 complex as well, and is simply aided by these homologous regions, since they have yet to be studied extensively. Next, we will cover real world evidence that makes me think this.
AMENDMENT: LET’S NOT FORGET THAT THE BRIDGING SHEET, AS COVERED IN THE OTHER AMENDMENT ABOVE, ALSO BINDS TO CCR5, AND IT IS PARTIALLY COMPOSED OF V LOOPS 1 AND 2. V LOOP 1 IS WHERE INSERT 3 LIES FROM THE PRADHAN PAPER, MEANING IT MAY HAVE PART OF THE BINDING BRIDGING SHEET THERE. ALSO, PRADHAN (AS PREVIOUSLY STATED) IS ASSESSING VARIANTS. I HAVE FOUND FOUR AMINO ACIDS IN INSERT 3, THAT CORRESPOND WITH WHAT SEEMS TO ME, TO BE THE ORIGINAL HIV V1 AND V2 REGIONS1415 (SYRL IN THE ORIGINAL V1 AND 2 REGIONS, AND RSLY IN SARS 2 GP 120 REGION. THIS IS THE SAME REGION THAT ALSO HAS THE ACIDS THAT MAY BIND TO CD4, THE MAIN RECEPTOR TARGET OF THE HIV GP120 COMPLEX (IMAGES BELOW)
AMENDMENT:
Original HIV sequence from above paper at the end of v loop 1 and 2 - TSYRL
Insert 3 from Pradhan, first 5 amino acids: RSYLT
Oh look, an amino acid V1 V2 chunk anagram. How coincidental. I think if anything is mimicking a bridging sheet, or any other interactive function normally facilitated by V loops 1 and 2, it's insert #3
First, evidence that the already covered mutation that defends against GP 120 binding in HIV, also protects in the event of acute SARS CoV 2 infection16:
“To date, two studies have evaluated a population-level correlation between CCR5-Δ32 mutation and COVID-19 prevalence and mortality, yielding contradictory results. Panda et al. (2020) explored 107 countries worldwide (data assessed on June 29, 2020) and reported a significant positive correlation between COVID-19 infection rate per million and mortality rate per million with the frequency of CCR5 Δ32 allele, as well as a positive correlation between CCR5-Δ32 allele frequency and COVID-19 mortality rate in an African population. Recently, we compared CCR5-Δ32 mutation frequency in 39 European countries with COVID-19 prevalence and mortality, as calculated on June 1, 2020, and found no association (Starčević Čizmarević et al. 2020). Both studies included data from the first COVID-19 wave, when most European countries had implemented restrictions on population movement to slow SARS-CoV-2 transmission and prevent health systems from becoming overwhelmed. At the end of June 2020, the EU re-opened internal borders, as did other European countries.
We were interested in whether the association of CCR5-Δ32 with COVID-19 prevalence and mortality in Europe remained unchanged during this second wave. Here, we present a statistical reanalysis of relevant data, performed on February 1, 2021, a year after the World Health Organization declared the outbreak to be a Public Health Emergency of International Concern. We obtained the prevalence of the CCR5-Δ32 allele in healthy individuals from 39 European countries based on Solloch et al. (2017). The analysis included the following countries: Albania, Austria, Belarus, Belgium, Bosnia and Herzegovina, Bulgaria, Croatia, the Czech Republic, Denmark, Estonia, Finland, France, Germany, Greece, Hungary, Iceland, Italy, Latvia, Lithuania, Luxembourg, Malta, Moldova, Montenegro, the Netherlands, North Macedonia, Norway, Poland, Portugal, Romania, Russia, Serbia, Slovakia, Slovenia, Spain, Sweden, Switzerland, Turkey, the United Kingdom, and Ukraine. Data for the prevalence (number of cases/106 inhabitants), mortality (number of deaths/106 inhabitants), number of diagnostic tests per 106 people, and time elapsed since the onset of the epidemic (days since January 1, 2020) in each country were obtained from a WorldOMeter website (www.worldometers.info/coronavirus/countries. Assessed February 1, 2021). We used multiple regression to evaluate the association between COVID-19 prevalence and mortality and CCR5-Δ32 prevalence, controlling for testing intensity and time elapsed since the onset of the epidemic in each country. To more closely approximate normal distributions for linear regression, we log transformed the data. Figure 1A and B show the correlation between COVID-19 prevalence and mortality and CCR5-Δ32 allele frequency in European populations.”
When assessed after the second wave, there was a clear negative correlation in 39 European countries with the CCR5-Δ32 mutation and the rate of both Covid cases and death, contradicting prior data, indicating it was protective, as it also is in HIV infection.
Early PCR diagnostics during the first “Covid wave” are notoriously inaccurate17. I would thus hypothesize, that population wide data concerning mass diagnostics predominately derived from PCRs and hastily concocted EUA antigen tests, would likely be more accurate for the second wave rather than the first, as quality went up and bad products were eliminated from use (and primers fixed of course). However outside the source I provided, I can't necessarily prove that point.
Here is another study concerning population data and the CCR5-Δ32 mutation's protective effects. This one is a much smaller and focused study that genotyped a portion of a Czech population during the first wave18
”In our study, we successfully genotyped 416 first-wave SARS-CoV-2-positive infection survivors (164 asymptomatic and 252 symptomatic) for CCR5Δ32, comparing them with a population based sample of 2,404 subjects. We found the highest number (P=0.03) of CCR5Δ32 carriers in SARS-CoV-2-positive/COVID-19-asymptomatic subjects (23.8 %) and the lowest number in SARS-CoV-2- positive/COVID-19-symptomatic patients (16.7 %), with frequency in the control population in the middle (21.0 %). We conclude that the CCR5Δ32 I/D polymorphism may have the potential to predict the severity of SARS-CoV-2 infection.”
“We found 21 % of the general population were carriers of at least one CCR5Δ32 allele. Representing a population allele frequency of 11.4 %, this finding is almost identical to that of a previous study by Drabek and Petrek (1998). They found 21.3 % of 386 Czech subjects were carriers of the CCR5Δ32 allele. Our data on genotype distribution and population frequency in the Czech population correspond with results from neighboring populations in Poland (Zwolińska et al. 2013) or Germany (Hütter et al. 2015). There were no differences between the entire group of SARS-CoV-2-positive subjects and population controls in respect of CCR5Δ32 deletion carriers (19.5 % vs. 21.0 %, P=0.48). Importantly, however, we observed a weak but significant trend in the frequency of CCR5Δ32 allele carriers (P=0.03; one-sided Cochran-Armitage trend test) between groups (Table 1). According to our expectations, the highest number of CCR5Δ32 carriers was found in SARS-CoV-2-positive/COVID-19-asymptomatic subjects (23.8 %) and the lowest in SARS-CoV-2-positive/COVID19-symptomatic patients (16.7 %), with frequency in the control population in the middle (21.0 %). How the CCR5Δ32 allele exactly protects against COVID-19 is not known. However, our results are in agreement with a summary of the indirect evidence (Mehlotra 2020), which indicates a promising new function of CCR5 beyond HIV-1/AIDS pathology”
“The CCR5Δ32 genetic variant has the potential to modify the CCR5-mediated organism response to many viral pathogens, including influenza and hepatitis viruses, cytomegalovirus as well as coronaviruses. While no functional receptor has been detected on cell surfaces in CCR5Δ32 homozygotes, reduced CCR5 expression has been observed in heterozygotes (Wu et al. 1997, Venkatesan et al. 2002). Therefore, it would appear this mutation acts as a natural model of complete CCR5 inhibition. Over-activation of the immune system, which is a hallmark of severe COVID-19 cases (Mehta et al. 2020, Paces et al. 2020), can be treated by CCR5 inhibition (Kliger and Levanon 2020, Pattersson et al. 2020). Mehlortra (2020) have summarized the results published thus far on the potential importance of Δ32 within CCR5, along with some other SNPs affecting CCR5 expression. They speculate that CCR5 inhibition may be of importance in COVID-19 treatment”
And so, once again, we find that the presence of this mutation which inhibits CCR5 binding from HIV GP120, is also protective in the event of SARS CoV 2 infection, to the point that it weakly correlates with asymptomatic infection in population data.
Next data point, CCR5 inhibition inhibits inflammatory cytokines and decreases SARS CoV 2 RNA in plasma19:
“Leronlimab, formerly known as PRO 140, is a CCR5-specific human IgG4 monoclonal antibody in development for HIV therapy as a once weekly, at home subcutaneous injection. In five completed and four ongoing HIV clinical trials where over 800 individuals received leronlimab, no drug-related deaths, serious injection site reactions, or drug–drug interactions were reported (Jacobson et al., 2008, 2010a; Jacobson et al., 2010b; Dhody et al., 2018). Subcutaneous, self-administration of leronlimab by patients facilitates simple, once weekly dosing. In contrast to the small molecule CCR5 inhibitors that prevent HIV Env binding to CCR5 via allosteric modulation, leronlimab binds to the CCR5 extracellular loop 2 domain and N-terminus, thereby directly blocking the binding of HIV Env to the CCR5 co-receptor via a competitive mechanism Leronlimab does not downregulate CCR5 surface expression or deplete CCR5-expressing cells, but it does prevent CCL5-induced calcium mobilization in CCR5+ cells with an IC50 of 45 mg/mL (Olson et al., 1999). This ability to specifically prevent CCL5-induced activation and chemotaxis of inflammatory CCR5+ macrophages and T cells suggests that leronlimab might be effective in mitigating pathologies involving the CCR5-ligand pathway”
At study day 0, all 10 critically ill patients received a subcutaneous injection of 700 mg leronlimab following baseline blood collection. Subsequently, the patients received a second subcutaneous injection of 700 mg leronlimab at study day 7. The defining features of severe COVID-19 disease include the plasma IL-6 and T cell lymphopenia (Huang et al., 2020; Lescure et al., 2020), so we longitudinally monitored these parameters for 2 weeks after the first leronlimab treatment. A reduction in the plasma IL-6 was observed as early as 3 days following leronlimab with a return to healthy control levels by day 14 (Figure 2A). In contrast to IL-6, the levels of other cytokines and chemokines were more variable after leronlimab treatment (Supplementary Figure 3). Following leronlimab administration, a marked restoration of CD8+ T cells (Figure 2B) and normalization of the CD4+ and CD8+ T cell ratio were observed in the blood samples (Figure 2C). These immunological changes occurred concomitant with leronlimab CCR5 receptor occupancy on the surface of CCR5+ T cells and monocytes (Figure 2D and E). The percentage of other CCR5-expressing cell types (CD4 T-cells, NK cells, and monocyte/macrophages) did not change significantly over 14 days of leronlimab treatment. Following leronlimab administration, SARS-CoV-2 plasma viremia decreased in all patients at day 7, and all but one patient had resolved SARS-CoV-2 plasma viremia to undetectable levels by day 14 (Figure 2F left, p = 0.0012). Finally, SARS-CoV-2 plasma viremia in leronlimab-treated COVID-19 patients was inversely correlated with the frequency of CD8+ T cells in the blood, which suggested immune restoration (Figure 2G).”
These critically ill SARS CoV 2 patients were given a monoclonal antibody that specifically binds to CCR5 at the height of their infections, one that is experimental for treating HIV infections. They noted a reversal of immune dysfunction (reduction of IL-6, rise of CD8 cells and a return to normal ratio of CD4/CD8) after application and CCR5 receptor occupancy by the antibody, resulting in a decrease of SAR CoV 2 plasma viral loads.
Let’s assess an alternate source that performed the same experiment20 (it should be noted that I do see repeating authors here however. Sources need vetting for potential conflicts of interest or instances of data dredging, to remain impartial). Here, we only get 4 critically ill patients. however once again, after application of the experimental monoclonal antibody and subsequent CCR5 receptor occupation, we see a reversal of immune dysfunction
“The novel coronavirus SARS-CoV-2 was identified in four critically ill patients who developed ARDS. Treatment target was the cytokine storm created by SARS-CoV-2 infection. After receiving leronlimab, all four patients initially survived. Two patients went on to recover and were discharged from hospital, while the other two patients subsequently died of surgical complications after making an initial recovery from SARSCoV-2 infection. All four patients clinically improved as measured by vasopressor support, and discontinuation of hemodialysis and mechanical ventilation. None of the patients developed a thromboembolic event after leronlimab injection.”
I have a single patient case study showing the same results21, but that may be beating a dead horse at this point. This concludes (for now) the gathering of evidence for CCR5 binding due to HIV GP120 homology
ADDITION:
It may be a while before I get to making the next article, so I’ll speed this process up a bit. Spike binding to CD4 T helper cells may, in my opinion, be a main culprit for both giant cell myocarditis, and giant cell arteritis. CD4 cells are considered the main culprit in giant cell myocarditis2223 and dendritic cells (remember DC sign?) and CD4 are the main culprits for giant cell arteritis24
And here are some examples of this occurring after SARS CoV 2 vaccinations2526.
UPDATE: THE FDA IS ACCUSING THE PEOPLE WHO CREATED AND ARE RESEARCHING THE DRUG IN THIS ARTICLE OF SECURITIES FRAUD, BASED ON HOW THEY TRIED TO ADVERTISE ITS EFFECTIVENESS. READ FOR YOURSELF TO DETERMINE HOW MUCH MERIT THE COUNTER CLAIMS HOLD
https://www.fda.gov/inspections-compliance-enforcement-and-criminal-investigations/warning-letters/cytodyn-inc-626957-02112022
ADDITION: INFORMATIVE VIDEO FOR STRONGER FUNDAMENTALS
https://en.wikipedia.org/wiki/T_helper_cell
https://en.wikipedia.org/wiki/Innate_immune_system
Comparative physiology of the pulmonary blood-gas barrier: the unique avian solution
American Journal of Physiology-Regulatory, Integrative and Comparative Physiology Vol. 297, No. 6
https://journals.physiology.org/doi/full/10.1152/ajpregu.00459.2009
The Effects of Secretory IgA in the Mucosal Immune System
Hindawi, BioMed Research International Volume 2020,
Article ID 2032057, 6 pages
https://doi.org/10.1155/2020/2032057
Microbiology and Molecular Biology Reviews
https://doi.org/10.1128/MMBR.05023-11
Detailed Dissection and Critical Evaluation of the Pfizer/BioNTech and Moderna mRNA Vaccines
MDPI
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8310186/pdf/vaccines-09-00734.pdf
Differences in Vaccine and SARS-CoV-2 Replication Derived mRNA: Implications for Cell Biology and Future Disease
OSFPREPRINTS (YET TO PASS PEER REVIEW, SUPPRESSION POSSIBLE, AUTHORS ARE WELL PUBLISHED AND KNOWLEDGEABLE)
https://osf.io/bcsa6/
Modeling of CCR5 Recognition by HIV-1 gp120: How the Viral Protein Exploits the Conformational Plasticity of the Coreceptor
MDPI
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8310383/pdf/viruses-13-01395.pdf
https://en.wikipedia.org/wiki/Chemokine
https://en.wikipedia.org/wiki/G_protein-coupled_receptor
Recent Advances targeting CCR5 for Cancer and its Role in Immuno-Oncology
Published in final edited form as: Cancer Res. 2019 October 01; 79(19): 4801–4807. doi:10.1158/0008-5472.CAN-19-1167.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6810651/pdf/nihms-1533705.pdf
A Conserved HIV gp120 Glycoprotein Structure Involved in Chemokine Receptor Binding
https://www.science.org/doi/10.1126/science.280.5371.1949
https://en.wikipedia.org/wiki/CCR5#CCR5-%CE%9432
HIV-1 Envelope Glycoprotein Variable Loops Are Indispensable for Envelope Structural Integrity and Virus Entry
PLOS ONE
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3731308/pdf/pone.0069789.pdf
Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01
10.1126/science.1192819
(Supp Fig 1.)
Could the CCR5-Δ32 Mutation be Protective in SARS-CoV-2 Infection?
Physiol. Res. 70 (Suppl. 2): S249-S252, 2021
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8884372/
Presence of mismatches between diagnostic PCR assays and coronavirus SARS-CoV-2 genome
Royal Society Open Science Published:10 June 2020
https://royalsocietypublishing.org/doi/10.1098/rsos.200636
CCR5Δ32 Deletion as a Protective Factor in Czech First-Wave COVID-19 Subjects
Physiol. Res. 70: 111-115, 2021
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8820511/pdf/pr70_111.pdf
CCR5 inhibition in critical COVID-19 patients decreases inflammatory cytokines, increases CD8 T-cells, and decreases SARS-CoV2 RNA in plasma by day 14
International Journal of Infectious Diseases
Article history: Received 16 October 2020 Received in revised form 27 October 2020 Accepted 30 October 2020
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7654230/pdf/main.pdf
Disruption of CCR5 signaling to treat COVID-19-associated cytokine storm: Case series of four critically ill patients treated with leronlimab
Journal of Translational Autoimmunity (4) 2021
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7823045/pdf/main.pdf
LERONLIMAB AND THE ROLE OF CCR5 SUPRESSION IN COVID-19 TREATMENT
CHEST INFECTIONS| VOLUME 160, ISSUE 4, SUPPLEMENT , A452, OCTOBER 01, 2021
https://doi.org/10.1016/j.chest.2021.07.445
IDIOPATHIC GIANT-CELL MYOCARDITIS — NATURAL HISTORY AND TREATMENT
The New England Journal of Medicine
https://www.nejm.org/doi/pdf/10.1056/NEJM199706263362603?articleTools=true
The CD4-gp120 interaction and AIDS pathogenesis
Annual Review of Immunology
Vol. 9:649-678 (Volume publication date April 1991)
https://www.annualreviews.org/doi/10.1146/annurev.iy.09.040191.003245?url_ver=Z39.88-2003&rfr_id=ori%3Arid%3Acrossref.org&rfr_dat=cr_pub++0pubmed
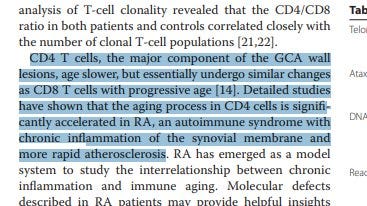
Giant cell arteritis: immune and vascular aging as disease risk factors
Mohan et al. Arthritis Research & Therapy 2011, 13:231
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3239337/pdf/ar3358.pdf
Biopsy-Proven Giant Cell Myocarditis Following the COVID-19 Vaccine
Circulation Heart Failure: Vol. 15, No. 4
https://www.ahajournals.org/doi/full/10.1161/CIRCHEARTFAILURE.121.009321
Giant cell myocarditis after first dose of BNT162b2 – a case report
European Journal of Heart Failure (2022) 24, 1319–1322
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9350328/pdf/EJHF-24-1319.pdf

















Completely OT: saw a comment from you on Unglossed and thought Rube Goldberg's razor...
Hmmmm Hanlon's razor = "Never attribute to malice that which is adequately explained by stupidity"
Rube Goldberg = "complicated gadgets performing simple tasks in indirect, convoluted ways"
Combine them and you have a substack name describing the pandemic: "Our 'leaders" designed complicated procedures, protocols and mandates to perform the simple task of keeping society 'safe' = 'healthy' via indirect, convoluted ways, not due to malice, but due to stupidity."
Stupidity mostly derived from "we gotta do something!!" and "polling shows the people we polled like what we're doing, keep doing it!!!", ignoring long-term debacle these actions will produce.
Perfect!
Alas it was Occam's razor, and invokes a more oxymoron combination - still clever, no question, but....
G'day. Nice to make your acquaintance.
Recently, I discovered this. https://github.com/lay295/TwitchDownloader/releases TwitchDownloader (for Windows, MacOS, Linux) that seems to work pretty good. Just past the video id (from the URL) into the VOD Link/ID box, click "Get Info" and if it finds it, it can pull it. There are sometimes errors, but when I closed the app and tried again in 30 minutes it worked well.
Anyhow, thought I'd share that before I forgot. Too bad there's no DMs in Substack (but there is a subscriber chat that the author can post to their subscribers).
I hope things are going well!